Welcome!
Welcome to the new CancerGRACE.org! Explore our fresh look and improved features—take a quick tour to see what’s new.
Have you ever wondered how exactly your oncologist knew what kind of cancer you had when he/she told you or a loved one about your diagnosis, whether it was non-small cell lung cancer (such as squamous cell or adenocarcinoma), breast cancer, or colon cancer?
Well, first there had to be a biopsy to obtain tissue from the tumor, which sometimes comes from a primary tumor (which is where the cancer originated) and sometimes from a metastatic site like the brain or bone. If the primary tumor is fairly obvious such as a big lung, colon, or breast mass, then most of the time we have a good idea what it is before we even get the pathology report. But it isn’t always immediately obvious where the primary site is located. Some patients come in with metastases to the brain, bone, lung, and/or liver, but with no obvious primary, and then we really rely on that biopsy to help us decide what type of cancer this is. Once the biopsy is obtained, it is sent to the pathology department.
Pathology, from the Greek pathologia, is the study of the underlying basis of disease, and pathologists are physicians who spend their days peering into microscopes looking at tissue samples trying to figure out what is abnormal about them. Once received, the biopsy tissue is fixed in something like formaldehyde (to keep the proteins and DNA from degrading when the tissue dies) and embedded in wax. The wax tissue blocks are then sliced into very thin slices and placed on microscope slides. Since most tissues are clear to the eye when sliced so thin, the tissue is stained with colored dyes to make the cells stand out under a microscope. Usually, the slides are also stained with antibodies (yes, the same kind that fight infection but these are mouse antibodies grown in a dish) that bind to specific proteins, and when combined properly can often separate a certain type of cell from another. Using these tools, pathologists can usually tell us an adenocarcinoma comes from the lung rather than from the colon or prostate, or they can differentiate a melanoma from mesothelioma from a sarcoma of the bone. But not always!
Sometimes we know something about a tumor, such as neuroendocrine or adenocarcinoma subtypes, but not where it originated. And all too often cancer is so poorly differentiated, or becomes so unlike the type of cell is originated from, that no one can tell much of anything about it at all. We have a name for these cases, called Cancers of Unknown Primary (or CUP). The reason this is an important topic is that some cancers are much more treatable than others, and some, such as germ cell cancer, are even curable if treated appropriately. In the future, hopefully all cancers will have effective treatments, so there will be a real need to know the type of cancer to tailor the treatment.
In truth, with the exception of monoclonal antibodies (developed in the 1980s), the tools that pathologists use are not all that different now than they were in the 19th century. Why are we relying on doctors peering into microscopes and going “Hmm… looks like lung to me”, when we live in the molecular era? We can test for EGFR and KRAS mutations, and send for other markers like bcr/abl and Her2 gene copy number using fluorescence in situ hybridization (FISH), so why don’t we have molecular tests to tell us what type of cancer is in that patient’s liver?
Well, my friends, that time is getting closer. There have been many attempts to classify types of cancer using molecular methods. Perhaps the most promising is something called oligonucleotide array technology. The genome comprises a person’s DNA, containing all 30,000+ so genes. When a cell wants to use a gene, say to signal itself to divide in response to a growth signal, is makes a copy of the DNA gene using something called messenger RNA (mRNA), which called a gene transcript. Kind of like a boss emailing off an order, the transcript then produces a desired effect. If you could analyze all of the thousands of emails (mRNAs) in a cell at one time (the transcriptome) you could essentially see everything that that cell was doing at the time, like a giant fingerprint or signature that would be totally unique to that cell or cancer type. Dr. West wrote a bit about this in 2007. For those who read the old post, array technology has gotten a bit more reliable and reproducible over the last 2 years, although it is still not perfect!
In a recently published study in the Journal of Clinical Oncology, investigators tried to design a way to use this molecular signature to tell the tissue of origin (TOO) of unknown tumor samples. They had previously developed a panel of 1,550 genes from 15 different TOOs using over 2000 tumors and normal tissues to get the best possible combination of genes that could distinguish one tissue from the others. They then tested this panel on 547 new tumor samples from the same 15 different tissues of origin (ranging from lung, breast, and liver to Hodgkin’s lymphoma). Half of the samples were from metastases, and half were from poorly differentiated tumors that were of known origin but were expected to look just like tumors that would otherwise be hard to tell just from looking under the microscope. They asked, can our test correctly tell us where these 547 samples originated from?
What they found was that the test was able to pick out the correct tumor type an impressive 87.8% of the time (ranging from 94% in breast to only 72% in pancreas). More impressively, if the tumor was NOT of a particular type, it correctly told you it was not that type 99.4% of the time (called specificity). So while the test couldn’t always say what you had, if it told you it was not lung cancer, it probably wasn’t!
This type of test would be really useful in the clinic if the results would always be this reliable, but is this ready for prime time? A major strength of this study was that the samples were collected, and the gene array data was collected, from 4 different centers using different microarray types, and that the data was all still comparable. Prior criticisms of array technology were that if you did the same experiment using the same array in 2 different labs, you got totally different answers, never mind using different array types! So this is a huge step forward to being able to use this technology routinely in your local hospital.
A continued problem with it, though, is the continued reliance on fresh frozen tissue to generate the signature. It is not always easy or even possible to get fresh tissue from a biopsy, and the processing of the mRNA is difficult, expensive, and time consuming. It will be much more useful if the test can be conducted on the wax-embedded tissue I mention above, and technically it should be possible. So I think while this is not yet ready for commercial production, it is very close, and then the poor pathologists will need to look for other things to occupy their time.
Imagine a day when technology like this can be applied to circulating tumor cells in the blood: a simple finger stick for a drop of blood, placed in a machine, could give you an accurate cancer diagnosis and generate a treatment plan in a few moments with none of the current fuss and uncertainties.
Hey, wait a minute: I would have to find something new to do, too!
Please feel free to offer comments and raise questions in our
discussion forums.
Hi app.92, Welcome to Grace. I'm sorry this is late getting to you. And more sorry your mum is going through this. It's possible this isn't a pancoast tumor even though...
A Brief Tornado. I love the analogy Dr. Antonoff gave us to describe her presentation. I felt it earlier too and am looking forward to going back for deeper dive.
Dr. Singhi's reprise on appropriate treatment, "Right patient, right time, right team".
While Dr. Ryckman described radiation oncology as "the perfect blend of nerd skills and empathy".
I hope any...
My understanding of ADCs is very basic. I plan to study Dr. Rous’ discussion to broaden that understanding.
Here's the webinar on YouTube. It begins with the agenda. Note the link is a playlist, which will be populated with shorts from the webinar on specific topics
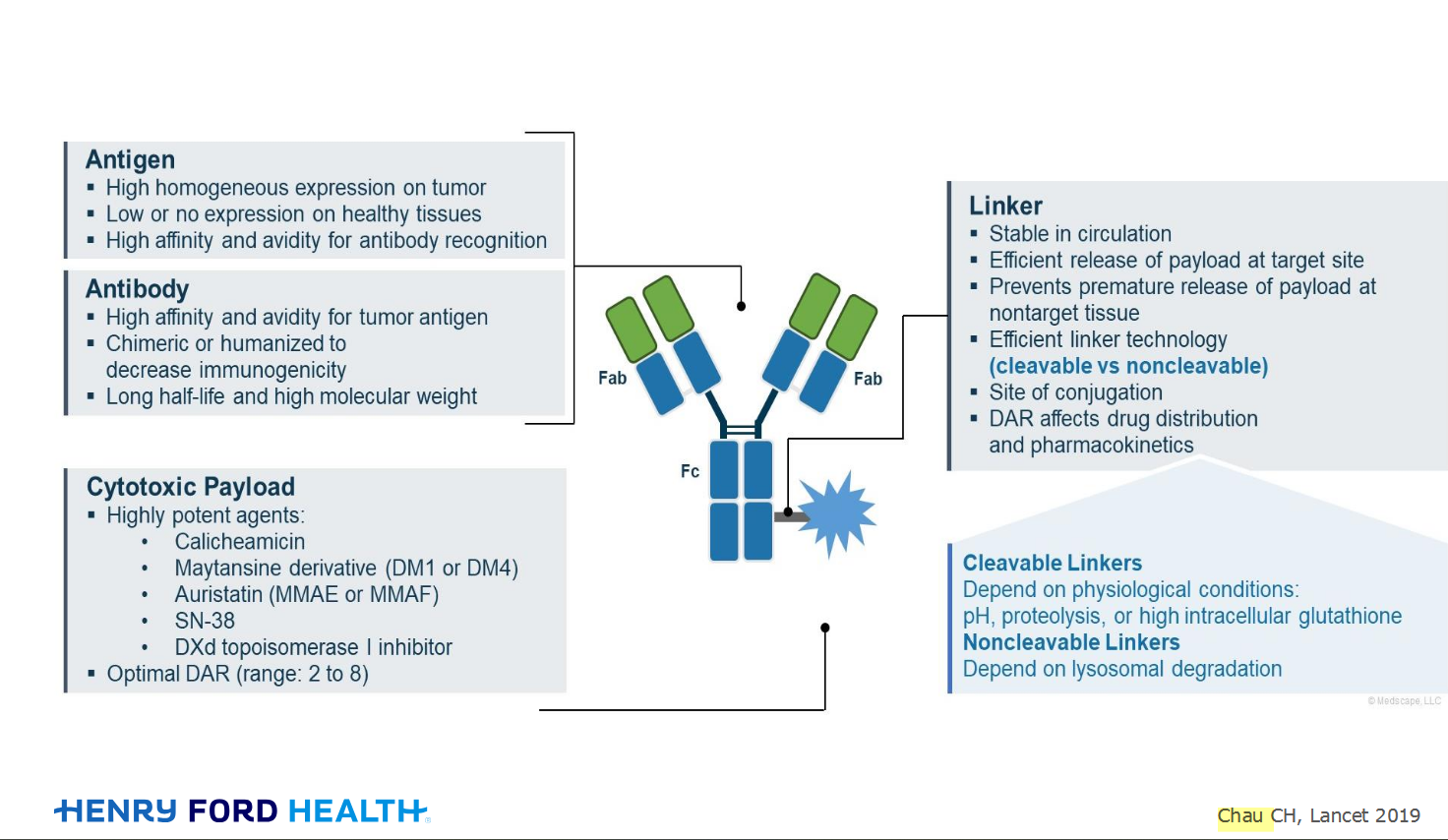
An antibody–drug conjugate (ADC) works a bit like a Trojan horse. It has three main components:

Welcome to the new CancerGRACE.org! Explore our fresh look and improved features—take a quick tour to see what’s new.