Welcome!
Welcome to the new CancerGRACE.org! Explore our fresh look and improved features—take a quick tour to see what’s new.
One of the questions that comes up fairly frequently is what to make of a "mixed response" to systemic therapy: after several weeks or months of treatment, a scan shows some areas of known disease shrinking, but others are growing. Why might this happen? What does it mean? And what should it lead us to do?
A recent report out of China looking at the heterogeneity of EGFR mutations in 180 patients who had paired sampling from different locations of their cancer (out of >3000 NSCLC patients who had EGFR molecular testing done at least once) helps provide a potential explanation, at least with regard to EGFR tyrosine kinase inhibitors (TKIs). These patients had either primary tumors detected at different times (called metachronous lesions), a primary cancer and lymph node pair, a primary cancer and distant metastasis biopsy pair, or paired samples from multiple lung nodules. The investigators found EGFR mutations in 119 primary tumors (50.6%), 15 (30.6%) lymph node metastases, 16 (45.7%) distant metastases, and 19 (46.3%) pulmonary nodules. And when looking at the matched samples from the same patient, they found that 25 of the 180 pairs (13.9%) were discordant. The discordance rate was 9-14% in all of the pairing combinations except for the situation of multiple lung nodules, in which case, the biopsies were discordant nearly a quarter of the time (10 of 41 cases, 24.4%). Patients with discordant pairs were the ones particularly likely to demonstrate a mixed response.
Not surprisingly, in patients who had molecular testing done from samples done at different times, the rate of discordance was lower in people who had received no systemic therapy (4 of 43, 9.3%) or chemotherapy only (8 of 59, 13.6%) than in people who had received an EGFR TKI therapy (10 of 38, 26.3%). This isn't surprising, since we might envision that the "selective pressure" that leads to cancer cells growing that don't have an EGFR mutation (or have an EGFR resistance mutation) when the cancer is otherwise effectively suppressed on an EGFR inhibitor. In fact, the paper cites a few cases of mixed response in which the primary tumor with an EGFR mutation was responding as a new lesion appeared and was found to not have an EGFR mutation. (We've seen a case in the GRACE community of someone whose EGFR mutated lung adenocarcinoma was responding to EGFR TKI therapy, as a new lesion appeared that was ultimately biopsied and found to be a squamous NSCLC without an EGFR mutation.)
These results are sobering as we try to guide our treatments by mutation results. First, a significant minority of the time, people will show different results depending on whether one area of a cancer is biopsied or another, and it appears that people with multiple pulmonary nodules are even more likely to have molecular heterogeneity from one nodule to the next. This provides a ready explanation of how treatment can lead to shrinkage in some lesions as others remain stable or grow, and/or new lesions appear. Why is the response rate to EGFR TKIs in patients with an EGFR mutation 60 or 70% and not 100%? Perhaps because many of the 30-40% who don't respond have growing lesions taht don't have an EGFR mutation, or the sample that showed a mutation was a minority of the overall cancer that doesn't truly represent most of the cancer's biology.
And while this principle is illustrated relatively clearly in patients with EGFR mutations, it's easy to translate these principles to conventional chemotherapy, which has its own molecular correlates that we don't understand as well. But we might well imagine that when we see some cancer lesions shrinking better than others, it's because the molecular features of the responding areas make them more susceptible to the effects of the chemotherapy than the resistant areas.
As we see these results, it reminds me of the patients I've seen who have responded very well to EGFR inhibitor therapy despite testing negative for a mutation, or even having a squamous cancer that we generally think of as minimally responsive to EGFR TKI therapy. Perhaps the sampled area just didn't represent the majority of the cancer's biology. These reports of heterogeneity remind me why I favor giving the vast majority of patients a trial of EGFR TKI therapy, whether their testing shows an EGFR mutation or wild type (no mutation). And of course, these data raise the specter that there may be patients who test negative for an ALK or ROS1 rearrangement due to a sampling error -- if a different area were tested, they'd get a positive result. For them, access to XALKORI (crizotinib) is tied directly to a positive test (at least for an ALK rearrangement, though still undefined for ROS1). While that's a very sad concept to consider, we can perhaps only be comforted by the idea that if someone has a molecular marker that is so difficulty to detect (perhaps it's represented in only a minority of the cancer), that person's cancer might not respond as well to targeted therapy. The report here can't answer that question.
It's also fair to ask what this implies about what to do. As I'd described in recent discussions of "acquired resistance" to EGFR TKIs or ALK inhibitor therapy, we'd expect that this is a molecularly heterogeneous state for many patients, as we often see limited progression against a background of good disease control. This work helps support the supposition that there are different molecular subpopulations whose proportions are reflected by the balance of progression vs. ongoing suppression/response. Just as many of the faculty here say that, in general, we view mixed responses as an individualized situation in which we weigh the degree of response vs. progression in our recommendation to switch treatment or continue on, we can use these findings to support the idea that if most of the cancer is still responsive to targeted therapy, it makes sense to continue the EGFR TKI or ALK inhibitor as we weigh the need to add local therapy or chemotherapy to address the growing portion: you're essentially treating the mutation-positive cancer with targeted therapy, and the mutation-negative cancer with another treatment strategy.
And so, yet again, we see evidence that more molecular testing, in this case more than one sample from the same patient, provides valuable insight, though at a cost of forcing us to redefine our over-simplified models, humbling us and our ability to explain and optimally treat people, at least until we return with newer and more complex models that might better reflect a more complex reality.
Please feel free to offer comments and raise questions in our
discussion forums.
Dr. Singhi's reprise on appropriate treatment, "Right patient, right time, right team".
While Dr. Ryckman described radiation oncology as "the perfect blend of nerd skills and empathy".
I hope any...
My understanding of ADCs is very basic. I plan to study Dr. Rous’ discussion to broaden that understanding.
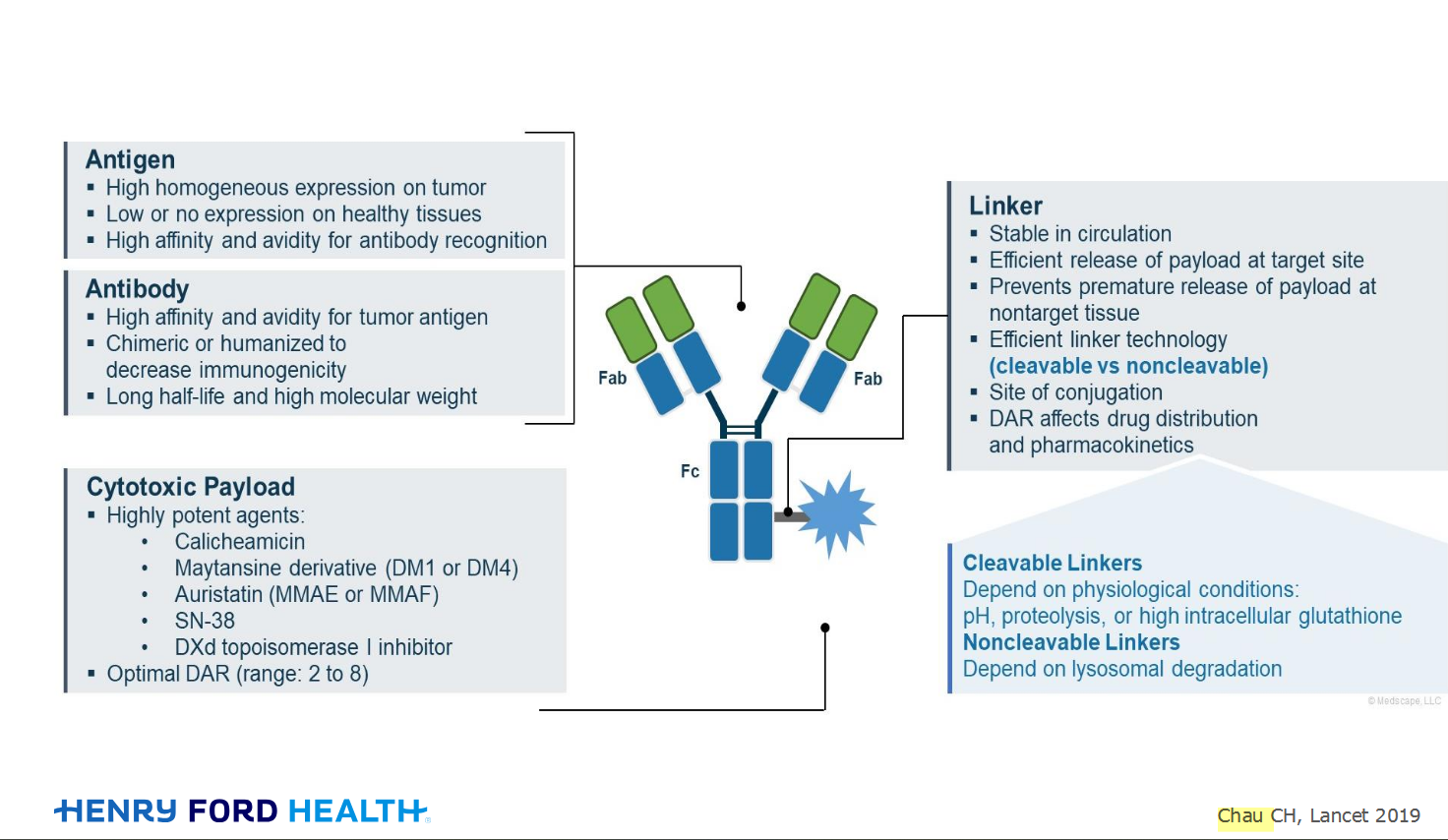
An antibody–drug conjugate (ADC) works a bit like a Trojan horse. It has three main components:
Bispecifics, or bispecific antibodies, are advanced immunotherapy drugs engineered to have two binding sites, allowing them to latch onto two different targets simultaneously, like a cancer cell and a T-cell, effectively...
The prefix “oligo–” means few. Oligometastatic (at diagnosis) Oligoprogression (during treatment)
There will be a discussion, “Studies in Oligometastatic NSCLC: Current Data and Definitions,” which will focus on what we...
Radiation therapy is primarily a localized treatment, meaning it precisely targets a specific tumor or area of the body, unlike systemic treatments (like chemotherapy) that affect the whole body.
The...

Welcome to the new CancerGRACE.org! Explore our fresh look and improved features—take a quick tour to see what’s new.
A Brief Tornado. I love the analogy Dr. Antonoff gave us to describe her presentation. I felt it earlier too and am looking forward to going back for deeper dive.