Welcome!
Welcome to the new CancerGRACE.org! Explore our fresh look and improved features—take a quick tour to see what’s new.
I’m just coming from a meeting on Targeted Therapies in the Treatment of Lung Cancer, which had the interesting format of dozens of 5-minute presentations just introducing or giving a very brief update of many new therapies. Some of these, such as the EGFR monoclonal antibody Erbitux (cetixumab), which I spoke on, have been approved by the US FDA for several years in other cancer settings and are well tested in humans. Others are so new that all that could be said about them is their chemical structure, presumed mechanism of action, description of how they killed some cancer cells in test tube models, and plans for moving ahead in very early trials in humans. I’ll describe some of the agents and classes in the next several weeks, with the caveat that some and perhaps many of these won’t be available for broad trials in lung cancer if they don’t get through several more hurdles.
But today I’m going to talk about the very interesting Keynote Lecture by Dr. Joseph Nevins of Duke Univeristy's Institute for Genome Sciences and Policy, on the topic of “Predictive Markers for Early Stage NSCLC”. As some of you may be aware from my prior post, other online information, or the general news, Duke has been a leading cancer center in studying genetic differences in lung cancer tumors, and it was investigators from there who recently published an important article in the New England Journal of Medicine on their ability to use genetic profiles to help predict outcomes for early, resected lung cancers. Dr. Nevins has been at the center of the efforts to individualize lung cancer care, and his lecture gave a broad overview of that work, essentially boiling down the questions he and his group are trying to answer as the following:
1) “who to treat”: which patients after surgery can be identified as having higher risk and requiring post-operative chemotherapy? which early stage patients can be identified as having a good enough prognosis to forego chemo?
2) “how to treat”: can gene arrays, the molecular signatures of a tumor, be employed to predict which particular chemotherapy drugs and/or targeted therapies would be most and least helpful for a particular lung cancer?
Dr. Nevins started by describing the work he and his lab have already been doing that was recently published in the New England Journal of Medicine (abstract here), evaluating resected stage IA tumors with the best overall prognosis based on a size less than 3 cm and no pleural or lymph node involvement. That’s as far as our current staging system can go, but we know that approximately 30% of stage I cancers recur and are all too often fatal. Moreover, stage IA cancers, despite still having some risk of recurrence and death, are considered to have too good a prognosis for to recommend adjuvant/post-operative chemotherapy in most cases. Obviously, it would be very helpful to identify those patients who cannot be identified by our current staging system as high risk but still have tumors that are aggressive and require more treatment. Dr. Nevins has been able to correlate genetic profiles of stage IA tumors with clinical outcomes for patients, leading to his group’s ability to discriminate a high-risk group that does remarkably better than the low-risk group, despite them all being classified as stage IA by the current system. Basically, it involves separating the patients with tumors that recurred from the patients who didn't have a recurrence, looking at the expression of lots of genes in all of them, and developing a "metagene" analysis of a series of genes that, depending on whether they have high or low expression, can reliably discriminate between the more and less aggressive cancers:
They then prospectively tested this model in patients at Duke and found that it worked well, and in fact far better than a clinical model that included patient age, sex, tumor size, and smoking status. Here are the survival curves for the different populations based on predictions from the metagene approach and a clinical model;
Looking at the data from the perspective of predicting how an individual patient would do, the metagene model was far more accurate in predicting which patients would recur within 2.5 years and which patients would not than the clinical model:
Here, the prediction of likelihood of recurrence based on the model is plotted vertically, so the more likely it is from the model that a patient will recur, the higher up the person's square will be. Also, the level of confidence in that prediction is reflected by the length of the bars above and below the squares -- short bars predict high confidence in the prediction, and wide bars demonstrate little confidence. The squares that are blue represent patients who did not recur, and the ones in red did recur. You can readily see that the genetic analysis did a remarkably better job of accurately predicting for a particular patient how likely recurrence was. And it could discriminate the aggressive from the less aggressive cancers with much greater confidence than the clinical model.
Encouraged that this approach works for the population of patients from Duke, the investigators then validated their results by predicting outcomes for patients with stage IA NSCLC tumors that were collected from trials at may other cancer centers as part of the Cancer and Leukemia Group B (CALGB), one of the major oncology cooperative groups in the US. Sure enough, the genetic profiles they had identified from their local tissue collection did an excellent job at predicting which patients would do especially well or poorly among stage IA patients.
But identifying higher risk or lower risk patients isn’t very helpful if you can’t do anything with that information. Remember that for stage IA patients, the general default recommendation after surgery is follow-up, observation but usually no chemotherapy or other treatment. So the trial that has been developed through the CALGB, designated Trial 30506, is a large randomized trial in which stage IA patients who undergo surgery will then have their tumor tissue characterized by gene expression analysis and then assigned a predicted outcome of Low Risk or High Risk. Low risk patients will go on to be observed according to standard practice, but high-risk patients will then be randomized to either be observed according to standard practice for stage IA NSCLC, or receive adjuvant chemotherapy on the basis of this new information. The schema is as shown in the figure here:
The study, led by Dr. David Harpole, an excellent thoracic surgeon at Duke, is in its final stages of approval and will likely be conducted over the next several years. It will directly evaluate whether survival is improved by this refinement of outcome prediction and treatment plans. Of note, this trial is only potentially adding treatment when it would otherwise not be recommended. An alternative use of this technology would be to use gene expression analysis to identify the lower risk stage IB, II, and IIIA patients who have undergone surgery and may not need chemo. However, right now we're a bit more comfortable suggesting a potential increase in treatment from the standard approach based on new information, rather than a potential decrease in treatment from the standard of care.
First things first. At least now you can get a good idea of how the science we're just developing now is shaping the clinical questions for today and may refine our standard treatment practices tomorrow. This is attempting to optimize our understanding of who to treat, and next we'll explore how to treat.
Please feel free to offer comments and raise questions in our
discussion forums.
Hi app.92, Welcome to Grace. I'm sorry this is late getting to you. And more sorry your mum is going through this. It's possible this isn't a pancoast tumor even though...
A Brief Tornado. I love the analogy Dr. Antonoff gave us to describe her presentation. I felt it earlier too and am looking forward to going back for deeper dive.
Dr. Singhi's reprise on appropriate treatment, "Right patient, right time, right team".
While Dr. Ryckman described radiation oncology as "the perfect blend of nerd skills and empathy".
I hope any...
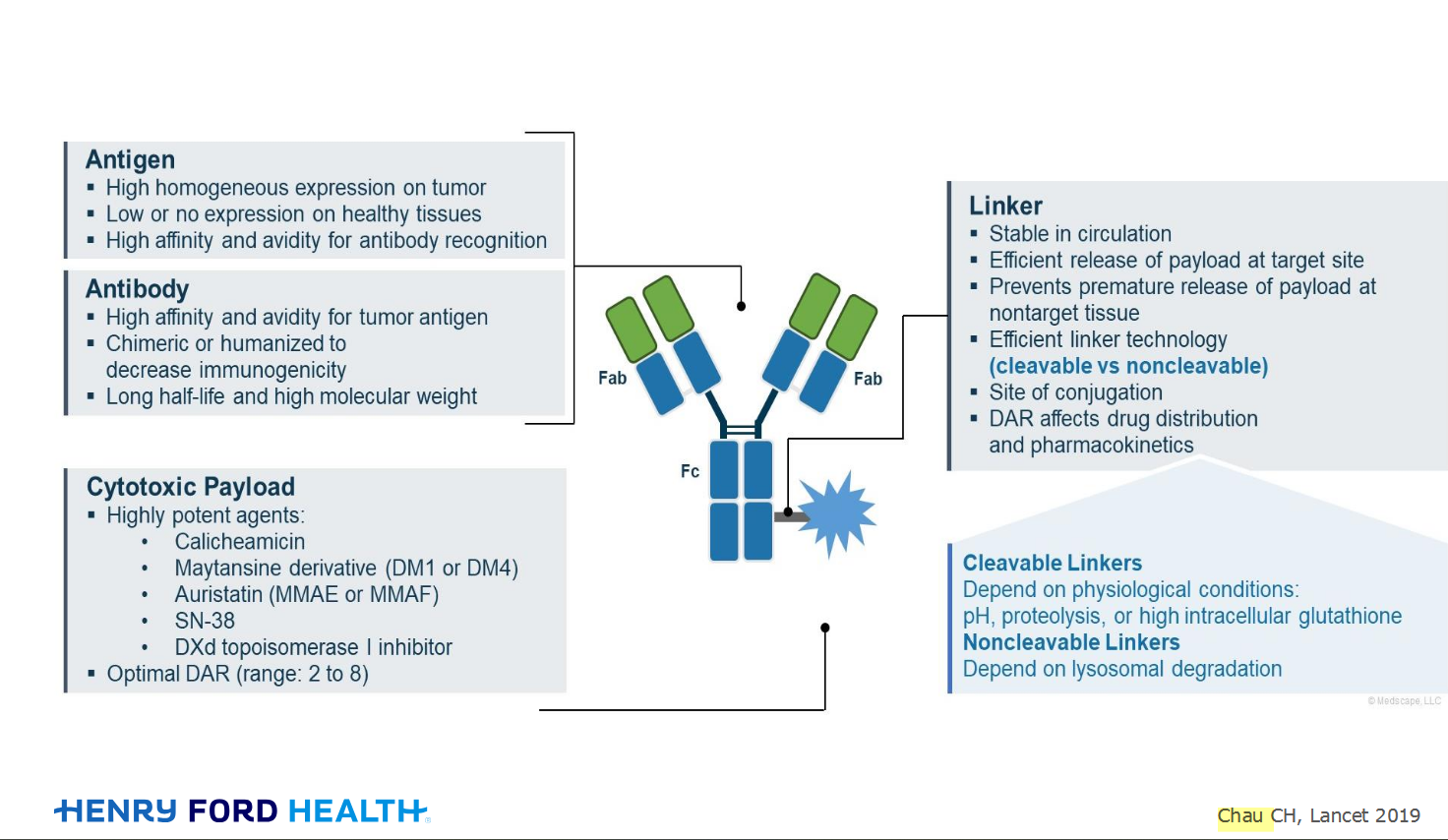
My understanding of ADCs is very basic. I plan to study Dr. Rous’ discussion to broaden that understanding.
Here's the webinar on YouTube. It begins with the agenda. Note the link is a playlist, which will be populated with shorts from the webinar on specific topics
An antibody–drug conjugate (ADC) works a bit like a Trojan horse. It has three main components:

Welcome to the new CancerGRACE.org! Explore our fresh look and improved features—take a quick tour to see what’s new.