Welcome!
Welcome to the new CancerGRACE.org! Explore our fresh look and improved features—take a quick tour to see what’s new.
In a recent issue of the New England Journal of Medicine, a research group from Massachusetts General Hospital in Boston published some very promising results from their work showing that they can now detect circulating tumor cells (CTCs) from most patients with lung cancer and even detect EGFR mutations and other molecular findings from these cells collected just from patient blood samples (abstract here).
This new approach is based on microfluidic technology, which fortunately isn’t necessary for you to understand but basically involves having blood flow through a closed, monitored space that detects proteins on the surface of tumor cells that aren’t present on other blood cells. These proteins are called EpCAMs, for Epithelial Cell Adhesion Molecules, and with this technique, tumor cells can be detected in varying quantities from patients with blood of patients with cancer, but these cells aren’t identified in the blood of patients without cancer (abstract here). But not only can these cells be detected and the numbers monitored, but the DNA within these cells can be assessed for DNA mutations such as activating mutations that predict response to EGFR tyrosine kinase inhibitors, and even other mutations that predict resistance to EGFR inhibitors, using some novel techniques of assessing DNA from individual cells and even “free DNA” floating in the plasma.
These investigators collected blood from a few dozen patients with advanced NSCLC, many with known EGFR mutations and some with the non-mutated (“wild-type”) EGFR gene. Some had received no prior therapy, while others had received prior chemo and/or EGFR inhibitors like tarceva (erlotinib) or iressa (gefitinib). They also assessed the correlation of numbers of CTCs in a patient’s blood with the tumor burden as measured by CT scan estimates. They found that they were able to detect CTCs in every cancer patient, but that the levels didn’t correlate well with the tumor burden on imaging. They mentioned that other factors might well be important, such as the invasiveness and the vascularity (density of blood vessels) of the cancer.
Although the numbers are small, the clear majority of patients with known EGFR activating mutations from samples of their tumor also were able to the same mutation detected from CTCs (11 of 12, 92%), which worked far better than detecting the mutation in free DNA of plasma (only 4 of 12, 33%). Detection of the marker of EGFR resistance, the so-called T790M mutation, was seen in several patients who had received and progressed on EGFR inhibitors (the presence of the T790M mutation is felt to be about 50% of the patients with resistance to EGFR inhibitors – so it’s not the only mechanism of resistance). Among patients who hadn’t received a prior EGFR inhibitor but had had a T790M mutation detected (which we’d predict would be associated with no benefit from subsequent EGFR inhibitor therapy), their survival was significantly lower than that seen in the patients who didn’t have a T790M mutation:
Interestingly, some of these patients actually had a response to EGFR therapy, but they were short-lived, so they tended to have disappointing results from EGFR inhibitors even when they did respond.
A few patients were able to have their blood drawn and have the CTC levels measured over time. In contrast with the lack of correlation between tumor burden and CTC numbers in an individual patient, changing CTC levels within the same patient over time were very well correlated with responses and progression on CT imaging. And by looking at EGFR mutations over time with new techniques, the investigators saw some new mutations developing over time, not just the T790M mutation but new activating mutations sometimes as time progressed.
All of this work has been done on just a few dozen patients at a single center, so this is far from routinely available standard practice. But it’s a remarkably important demonstration for what is possible. Specifically, it’s now possible to detect and get genetic information from cancer cells collected from blood samples, and that changing numbers of these cells over time may give us information about how a patient is responding to treatment, in between or in addition to imaging results. This is particularly important because, as I had just noted in my response to nlrohde about collecting tissue from lung cancer patients, the lack of accessibility of tumor tissue from lung cancer patients is a limitation in our understanding of lung cancer, compared to the greater ability to study cancer cells from patients with leukemia, melanoma, or even breast cancer, for whom getting tissue may be an invasive procedure but doesn’t usually involve going into the chest cavity. I believe that being able to access cancer cells with a blood draw, and potentially to use these cells to monitor the status of the cancer over time, is likely to be a true breakthrough.
Please feel free to offer comments and raise questions in our
discussion forums.
Dr. Singhi's reprise on appropriate treatment, "Right patient, right time, right team".
While Dr. Ryckman described radiation oncology as "the perfect blend of nerd skills and empathy".
I hope any...
My understanding of ADCs is very basic. I plan to study Dr. Rous’ discussion to broaden that understanding.
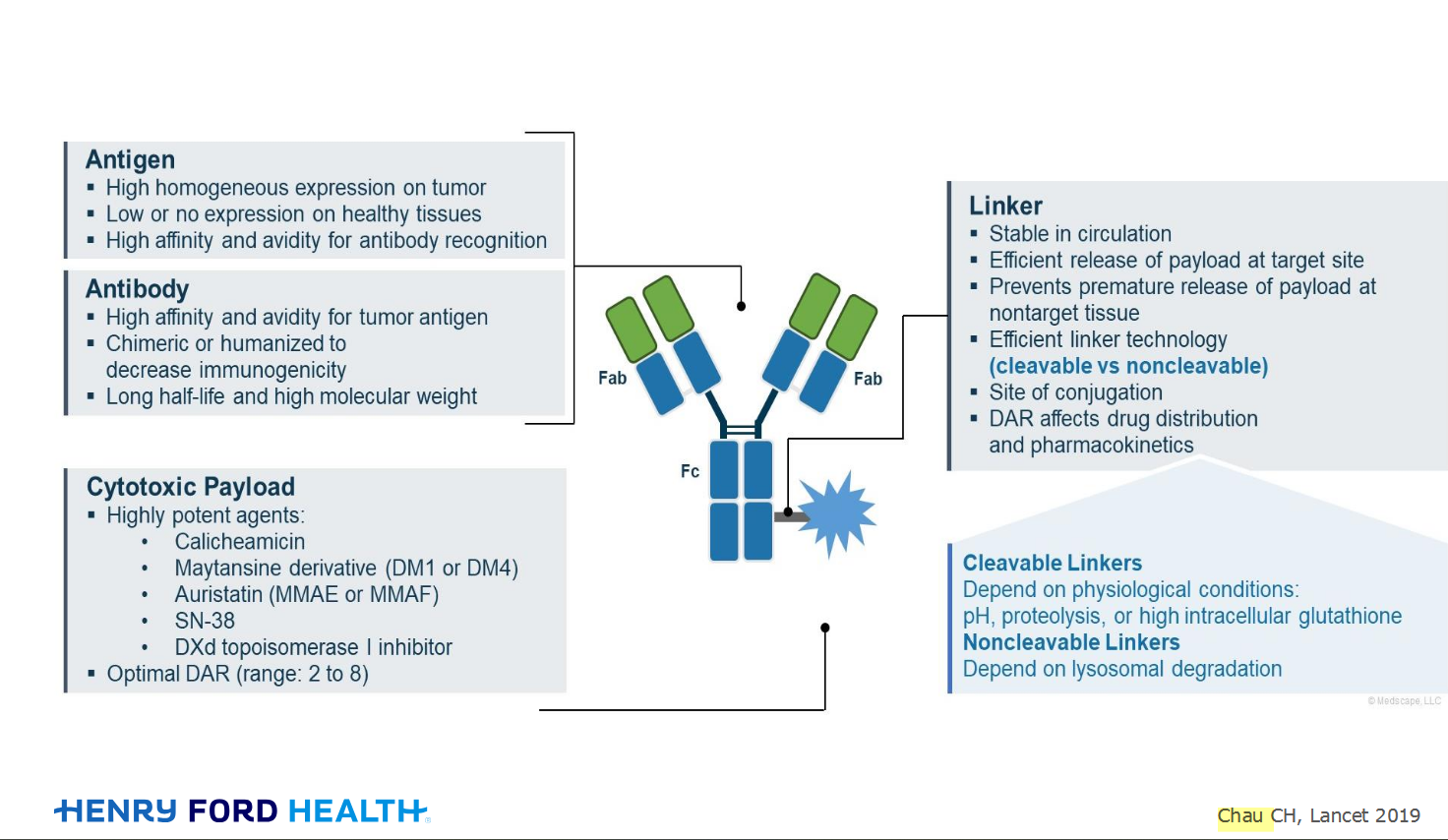
An antibody–drug conjugate (ADC) works a bit like a Trojan horse. It has three main components:
Bispecifics, or bispecific antibodies, are advanced immunotherapy drugs engineered to have two binding sites, allowing them to latch onto two different targets simultaneously, like a cancer cell and a T-cell, effectively...
The prefix “oligo–” means few. Oligometastatic (at diagnosis) Oligoprogression (during treatment)
There will be a discussion, “Studies in Oligometastatic NSCLC: Current Data and Definitions,” which will focus on what we...
Radiation therapy is primarily a localized treatment, meaning it precisely targets a specific tumor or area of the body, unlike systemic treatments (like chemotherapy) that affect the whole body.
The...

Welcome to the new CancerGRACE.org! Explore our fresh look and improved features—take a quick tour to see what’s new.
A Brief Tornado. I love the analogy Dr. Antonoff gave us to describe her presentation. I felt it earlier too and am looking forward to going back for deeper dive.