Welcome!
Welcome to the new CancerGRACE.org! Explore our fresh look and improved features—take a quick tour to see what’s new.
Continuing on the subject from my last post of gene signatures to predict clinical behavior, another ASCO presentation came from Poland (abstract here), with the goal of validating a couple of different kinds of gene signatures that have been presented before. One, from a group in Taiwan, was with just 5 genes and had been published in the New England Journal of Medicine (abstract here), while another with just 3 genes had been reported at a prior ASCO meeting (abstract here) by the same Polish group that was presenting this year. They had snap frozen tissue available from 142 patients in Poland, ranging in stage from IA to IV, although most were from earlier stage patients. They pooled results across all of the stages, though, which isn’t a very pure way to interpret the data.
The first thing that they showed was that the three gene signature was able to discriminate a group with better vs. worse prognosis when both adenocarcinoma and squamous cell NSCLC patients were pooled together, but the five-gene signature showed a more modest difference that wasn’t statistically significant:
However, both gene signature approaches were able to do a better job when they were only trying to compare adenocarcinoma patients or squamous cell patients to each other:
I was assigned the task of providing expert commentary on this abstract among several others in the setting of early stage NSCLC at ASCO. As I mention in my summary slide below, my feeling was that this microarray work does have some ability to discriminate better prognosis from worse prognosis patients, but the curves don’t separate so strikingly that I really think you could or should make treatment decisions, such as whether to recommend adjuvant chemotherapy, based on these results. Overall, it’s not clear to me that this technically challenging work is really better at discriminating high risk vs. low risk patients better than tumor grade, tumor uptake (SUV) on PET scan, or any of a number of other readily available factors. You can contrast the modestly separated curves above with the huge difference in outcomes between high risk and low risk groups defined by using a different approach I had previously described, the “metagene” approach pioneered at Duke (shown in slide below).
I also noted that the pooling of patient results from stage I all the way to stage IV was on the sloppy side.
But the greatest limitations are from two issues. The first is that this work requires tissue to be collected and frozen at the time of surgery in a way that requires some extra time, equipment, and extra attention. Right now, only a few of the most research-oriented centers do this regularly, so it would require a change of practice. Secondly, there are currently a number of different gene signatures out there, each competing to become a useful tool, but none with a critical mass of data to support it. Thankfully, this work is about to enter into larger, multi-institutional trials, such as one being launched by the cancer cooperative research group CALGB that will attempt to use the Duke metagene model to refine which stage I patients should or should not receive adjuvant chemotherapy. This work will be very important to see whether adequate tissue for analysis can be completed in the broader community and whether the very impressive results with gene signature approaches that we’ve seen in small single-institution studies can be replicated on a much larger scale.
Please feel free to offer comments and raise questions in our
discussion forums.
Hi app.92, Welcome to Grace. I'm sorry this is late getting to you. And more sorry your mum is going through this. It's possible this isn't a pancoast tumor even though...
A Brief Tornado. I love the analogy Dr. Antonoff gave us to describe her presentation. I felt it earlier too and am looking forward to going back for deeper dive.
Dr. Singhi's reprise on appropriate treatment, "Right patient, right time, right team".
While Dr. Ryckman described radiation oncology as "the perfect blend of nerd skills and empathy".
I hope any...
My understanding of ADCs is very basic. I plan to study Dr. Rous’ discussion to broaden that understanding.
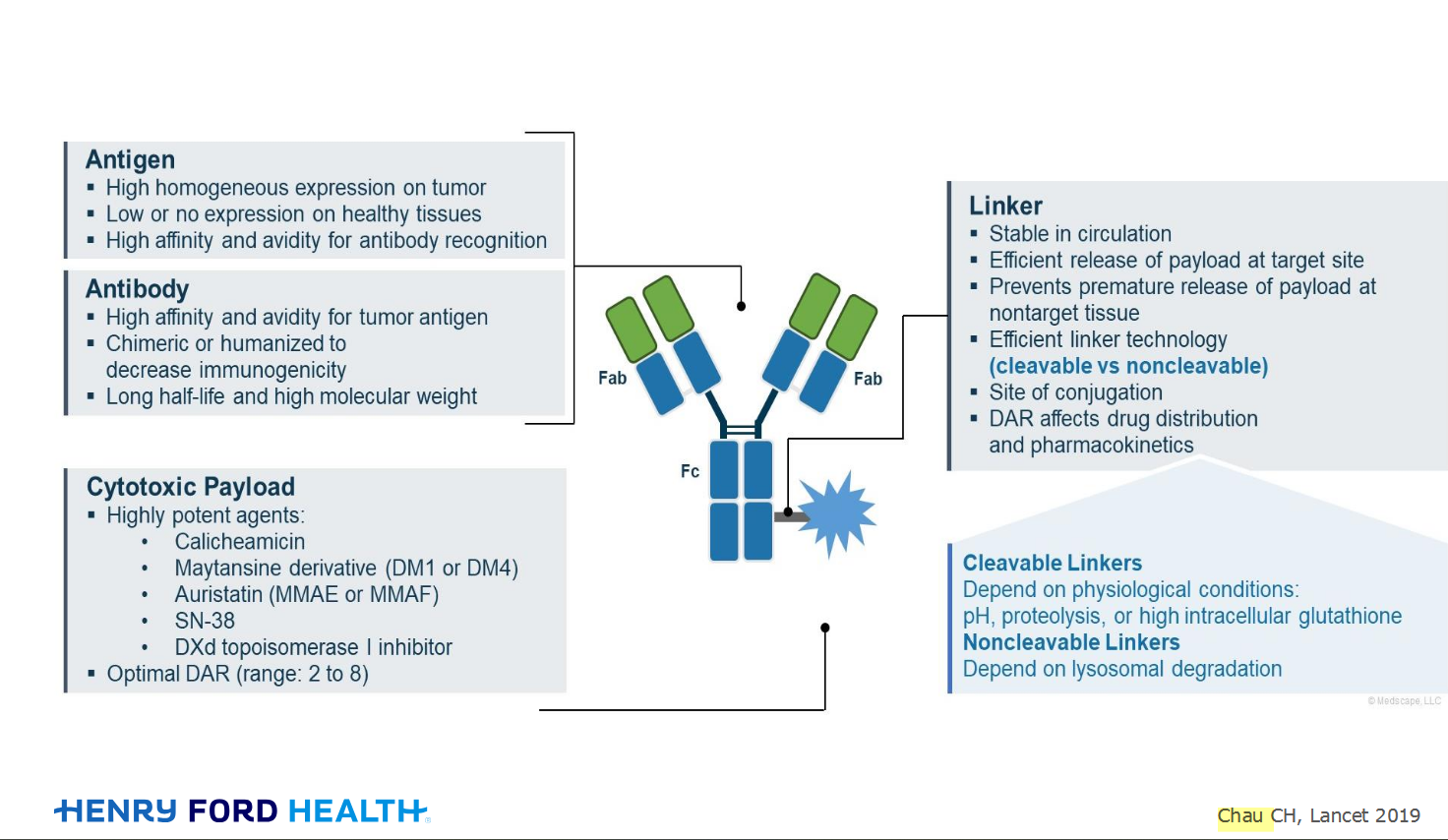
Here's the webinar on YouTube. It begins with the agenda. Note the link is a playlist, which will be populated with shorts from the webinar on specific topics
An antibody–drug conjugate (ADC) works a bit like a Trojan horse. It has three main components:

Welcome to the new CancerGRACE.org! Explore our fresh look and improved features—take a quick tour to see what’s new.