Welcome!
Welcome to the new CancerGRACE.org! Explore our fresh look and improved features—take a quick tour to see what’s new.
NOTE: ALL FIGURES CAN BE SEEN BY DOUBLE-CLICKING ON THEM, EVEN THOUGH NOT ALL APPEAR AS THUMBNAIL VIEWS PROPERLY.
We've discussed various ways of predicting outcomes with EGFR inhibitors like Tarceva or Iressa using clinical variables like smoking status or BAC subtype, as well as molecular markers like EGFR mutations, or EGFR gene amplification or protein expression. These can all be of value, but we know that the clinical markers are quite inexact, while the molecular markers are still a work in progress. Moreover, the molecular testing that has been the subject of most of the work thus far has come from tumor tissue material, which is often hard to come by and usually requires a biopsy or resection to obtain. But a recent article in the Journal of the National Cancer Institute, by a international collaborative group led by Dr. David Carbone from Vanderbilt University, describes their recent success in predicting survival after administration of EGFR inhibitor therapy using serum samples from patients all around the world (abstract here -- full article also available from that page).
Dr. Carbone and his team at Vanderbilt have been leaders in the field of serum proteomics, which is the study of the proteins in the serum, the straw-colored fluid that remains after blood has clotted, so the cells and clotting proteins are absent. Obviously, collecting blood, from which serum samples can be analyzed, is much easier than collecting extra cancer cells in a biopsy to send off for studies. The approach they used at Vanderbilt is called matrix-assisted laser desorption ionization (MALDI) mass spectroscopy (MS), or just "mass spec", which is very complex (code, perhaps, for me saying I really don't understand it well? we only had to do a year of college physics to get into medical school, you know), but basically it is a way of analyzing a serum sample to report a set of peaks that represent different proteins in the sample:
You can use computer software to analyze the peak patterns of a large collection of serum samples from cancer patients before treatment, tell the software how those patients did, and then have the complex software discern patterns that can discriminate the patients who do well from the ones who do poorly. We saw a similar approach to this, done with tumor tissue, from the lab of Dr. Nevins at Duke, described in a prior post. For the study of serum, Taguchi and colleagues (including Dr. Carbone and many other stars in lung cancer from other institutions) started with a training set of 139 samples from patients in Italy and Japan who received Iressa after the serum was collected. From there the software was able to discern a set of eight peaks that could separate the group destined to do well from those who subsequently did poorly. The invstigators then tested their model with patient samples that served as validation sets to see if their model developed from the training set held up. One group was 67 more patients from Italy who received Iressa, and a second group was 96 patients from the US-based Eastern Cooperative Oncology Group (ECOG), from a trial called ECOG 3503 that gave Tarceva as first-line therapy for patients with advanced NSCLC.
It worked. Looking at the Italian patient that received Iressa, the group that the analysis predicted as good outcome had a median progression free survival of 84 days vs. 61 days for the poor outcome group, and the median overall survival difference was 207 vs. 92 days. In the ECOG set, the survival difference was a striking 306 vs. 107 days for good and poor outcome groups, respectively:
These survival curves are flanked by hatched lines that are the "confidence intervals" that are wider if there's a lot of variability in the data, and they're tighter around the solid line when there's less varibility in the results. I'd focus more on the differences in the solid lines.
Interestingly, the investigators also showed that their analysis could discriminate groups that would do better or worse even among populations that we generally don't think of as doing as well with EGFR inhibitors. Here's the convincing differences in the survival curves for the smokers in the Italian validation set:
Now, you may be thinking, "Big deal...you can predict who is going to live longer with this fancy mass spectroscopy stuff, but I can do that by seeing who's active and who's sleeping all day, or who's lost 30 pounds before starting treatment." OK, it's true that there are many ways to estimate prognosis, which is to a good degree a function of the health of the patient. But the investigators checked their technique to see if it predicted outcomes in patients who received surgery for early stage disease, or chemo and not EGFR inhibitors for later stage disease. And the mass spec patterns didn't predict outcome very well. Instead, this technique was predictive of how people would do with EGFR inhibitors like Iressa or Tarceva, but this was specific for these treatments, rather than just being another prognostic factor that indicates which patients will do better or worse than other patients no matter what therapy they receive.
This approach is still only tested in relatively small numbers of patients. However, the investigators did the analysis in two different labs, one at Vanderbilt and another at the University of Colorado, and they both produced very similar, reproducible results for what the peak signature would look like whether samples were run in Denver or Nashville. While caveats were raised that sometimes early proteomics work has not panned out after initial promise in single institutions, and that the surgery and chemo recipients in their "control population" were a different patient mix than were enrolled on the Iressa and Tarceva trials they focused on, an accompanying editorial by the people who ran the influential BR.21 trial (editorial here) noted that this work was very encouraging.
In fact, there is a company called Biodesix that is now marketing this technology. You can have your serum run at a local lab and have the data analyzed through there company, or have a small amount of your serum sent to their lab in Boulder, Colorado for them to run the analysis and interpretation. Sample report information is available here (it's pretty dense and boring when all you want to know is "good" or "poor"). The pricing isn't listed, and I haven't called yet to find out pricing. The authors of the JNCI paper note that it's a great advantage to have a reliable technique that can work on just a tiny amount of serum and "at low cost", but we'll have to see how low cost it really is as a commercial test.
This work hasn't yet emerged as the definitive answer, and it's certainly not a standard approach at this time. Nevertheless, this is predictive work based on well conducted research that has been published after careful review, as part of a worldwide collaboration of clinical and lab researchers. Several very prominent experts are part of the company's advisory team, so this work is certainly poised to potentially break into mainstream, widespread use. But I'll come back to the question of whether people would really want to know that a treatment will or won't work. It's great if you learn you're likely to benefit, but would you rather learn that you are highly unlikely to benefit from a treatment, or would you rather have some unknowns out there and have more hope? What if insurers said that they're only going to cover the subset who are in the good group, and the others need to pay for it themselves because it's futile therapy? I think this is the future of oncology, and that it will represent a great advance when we can utilize the drugs we already have much more intelligently. But it should lead us to predict that some people are unlikely to benefit from any treatment, and they may prefer the hope that exists with the unknown over new information that suggests such hope is misguided.
I welcome your thoughts. I'm also going to re-open an older poll that covers some of the issues raised here.
Please feel free to offer comments and raise questions in our
discussion forums.
A Brief Tornado. I love the analogy Dr. Antonoff gave us to describe her presentation. I felt it earlier too and am looking forward to going back for deeper dive.
Dr. Singhi's reprise on appropriate treatment, "Right patient, right time, right team".
While Dr. Ryckman described radiation oncology as "the perfect blend of nerd skills and empathy".
I hope any...
My understanding of ADCs is very basic. I plan to study Dr. Rous’ discussion to broaden that understanding.
Here's the webinar on YouTube. It begins with the agenda. Note the link is a playlist, which will be populated with shorts from the webinar on specific topics
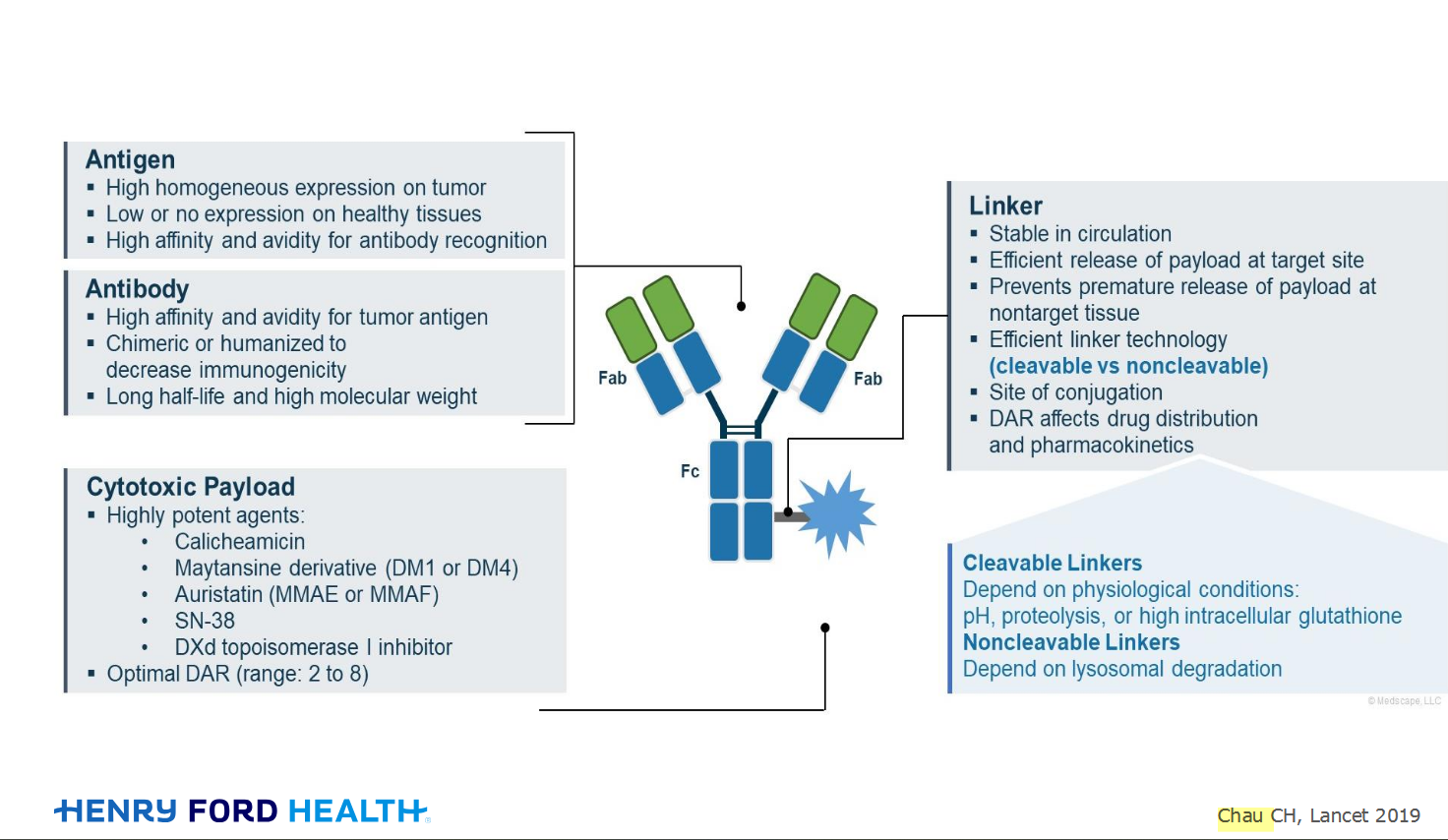
An antibody–drug conjugate (ADC) works a bit like a Trojan horse. It has three main components:
Bispecifics, or bispecific antibodies, are advanced immunotherapy drugs engineered to have two binding sites, allowing them to latch onto two different targets simultaneously, like a cancer cell and a T-cell, effectively...

Welcome to the new CancerGRACE.org! Explore our fresh look and improved features—take a quick tour to see what’s new.
Hi app.92, Welcome to Grace. I'm sorry this is late getting to you. And more sorry your mum is going through this. It's possible this isn't a pancoast tumor even though...